For this example, we’ll explore the curve_lik() function, which can help generate profile likelihood functions, and deviance functions with the help of the ProfileLikelihood package.1. For an introduction to what profile likelihoods are, see the following paper.2
library(ProfileLikelihood) #> Loading required package: nlme #> Loading required package: MASS
We’ll use a simple example taken directly from the ProfileLikelihood documentation where we’ll calculate the likelihoods from a glm model
data(dataglm) xx <- profilelike.glm(y ~ x1 + x2, data = dataglm, profile.theta = "group", family = binomial(link = "logit"), length = 500, round = 2 ) #> Warning message: provide lo.theta and hi.theta
Then, we’ll use curve_lik() on the object that the ProfileLikelihood package produced.
lik <- curve_lik(xx, data = dataglm)
Next, we’ll plot four functions, the relative likelihood, the log-likelihood, the likelihood, and the deviance function.
ggcurve(lik[[1]], type = "l1", nullvalue = TRUE)
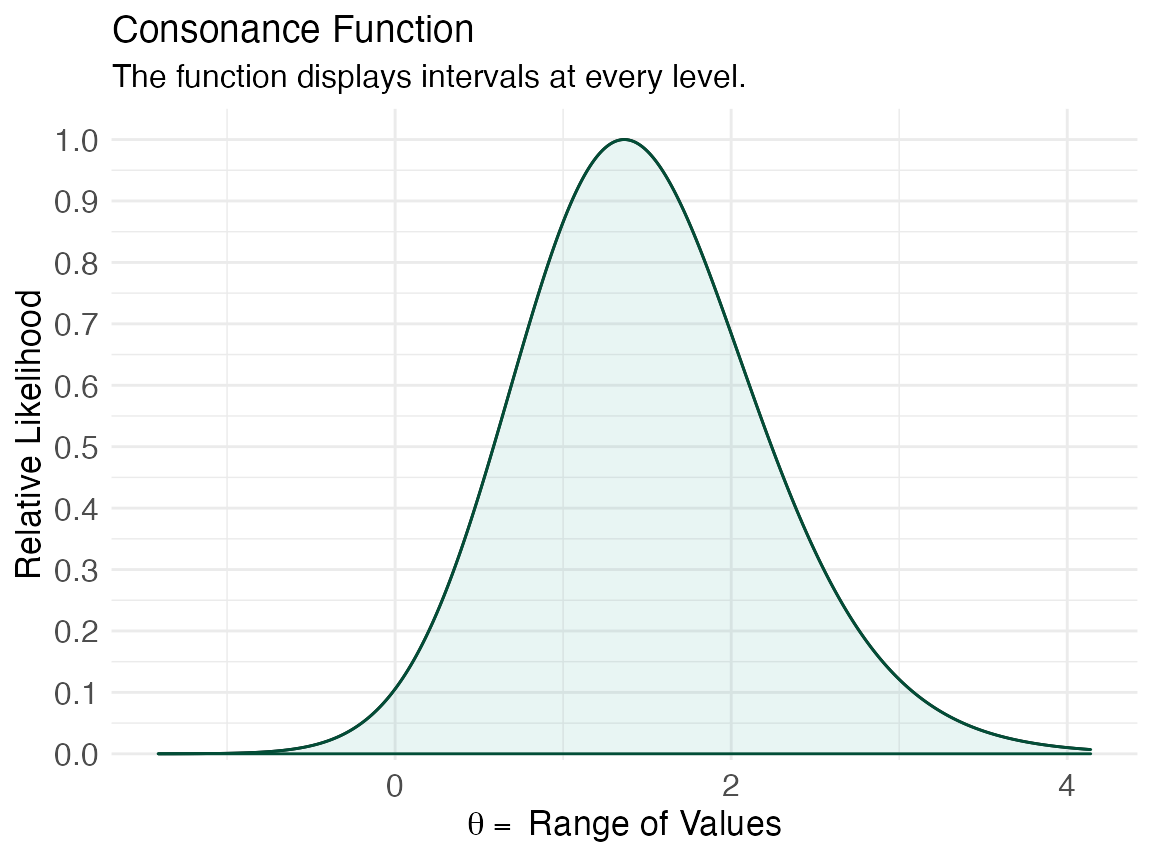
ggcurve(lik[[1]], type = "l2")
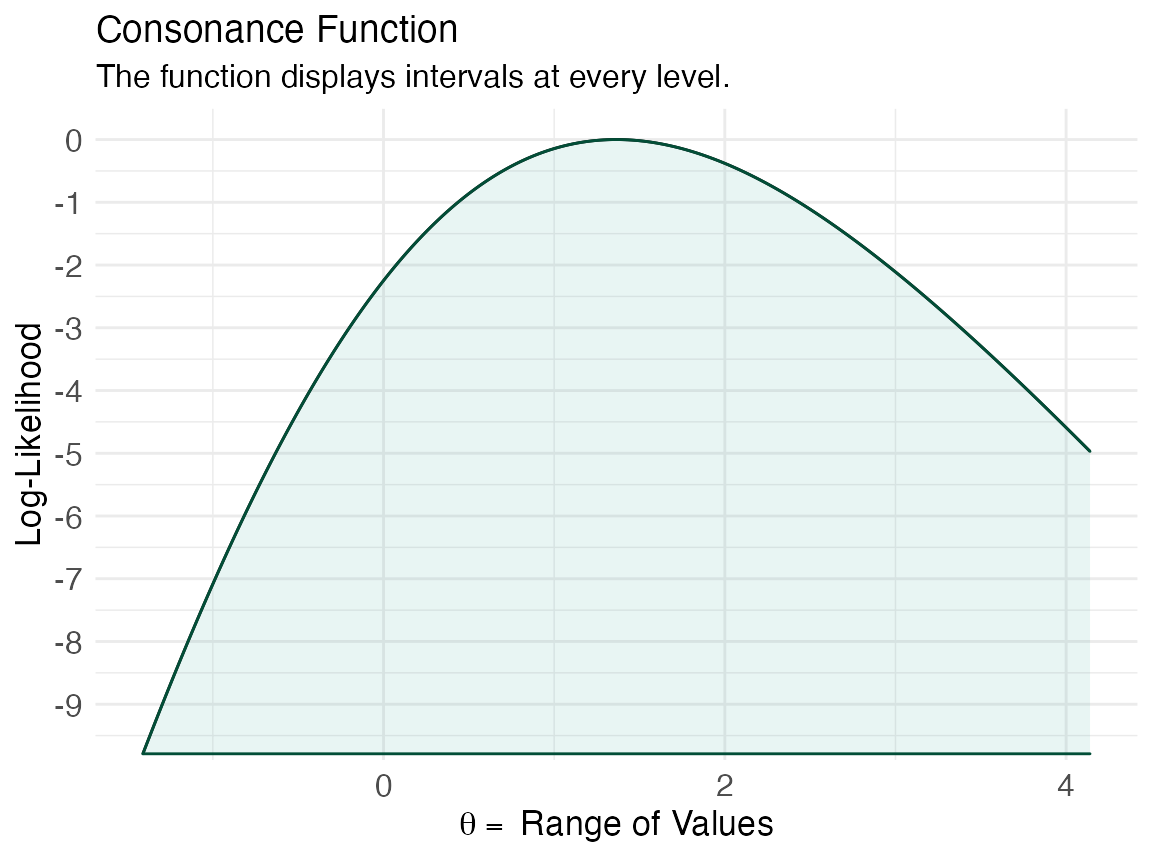
ggcurve(lik[[1]], type = "l3")
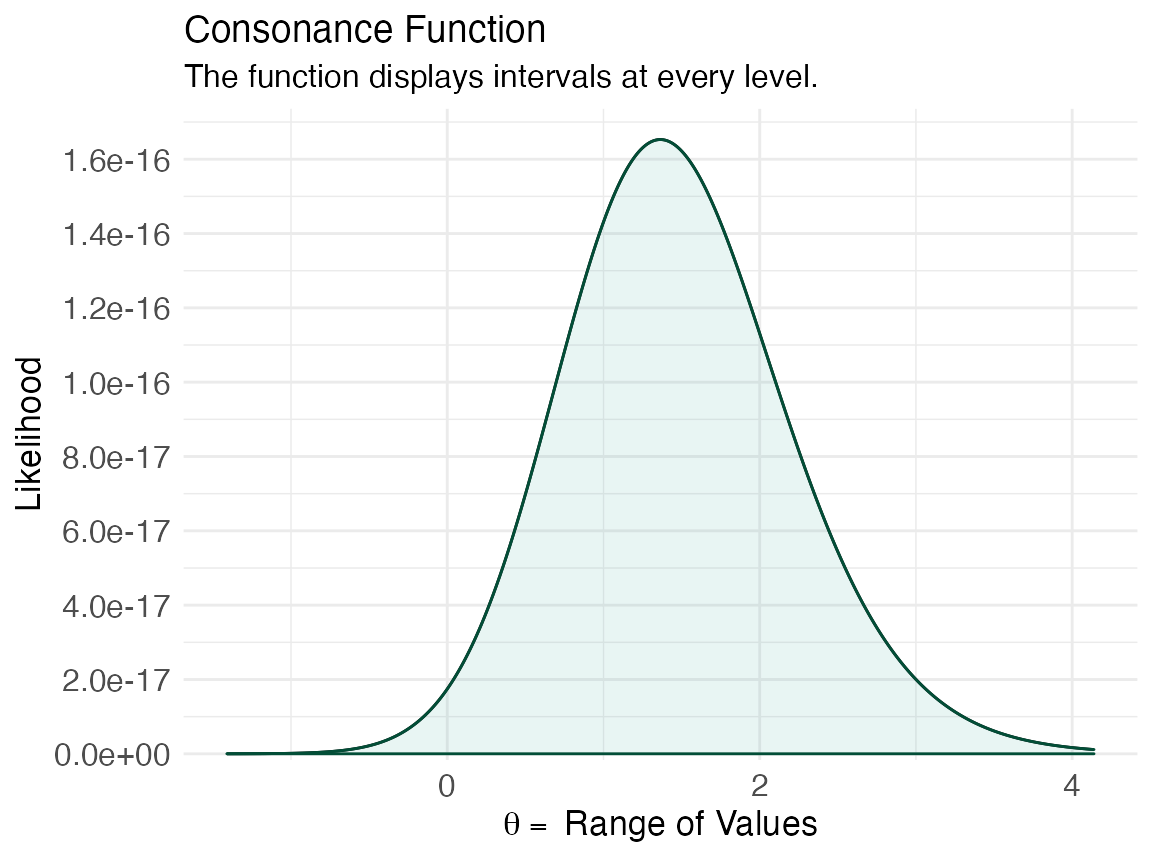
ggcurve(lik[[1]], type = "d")
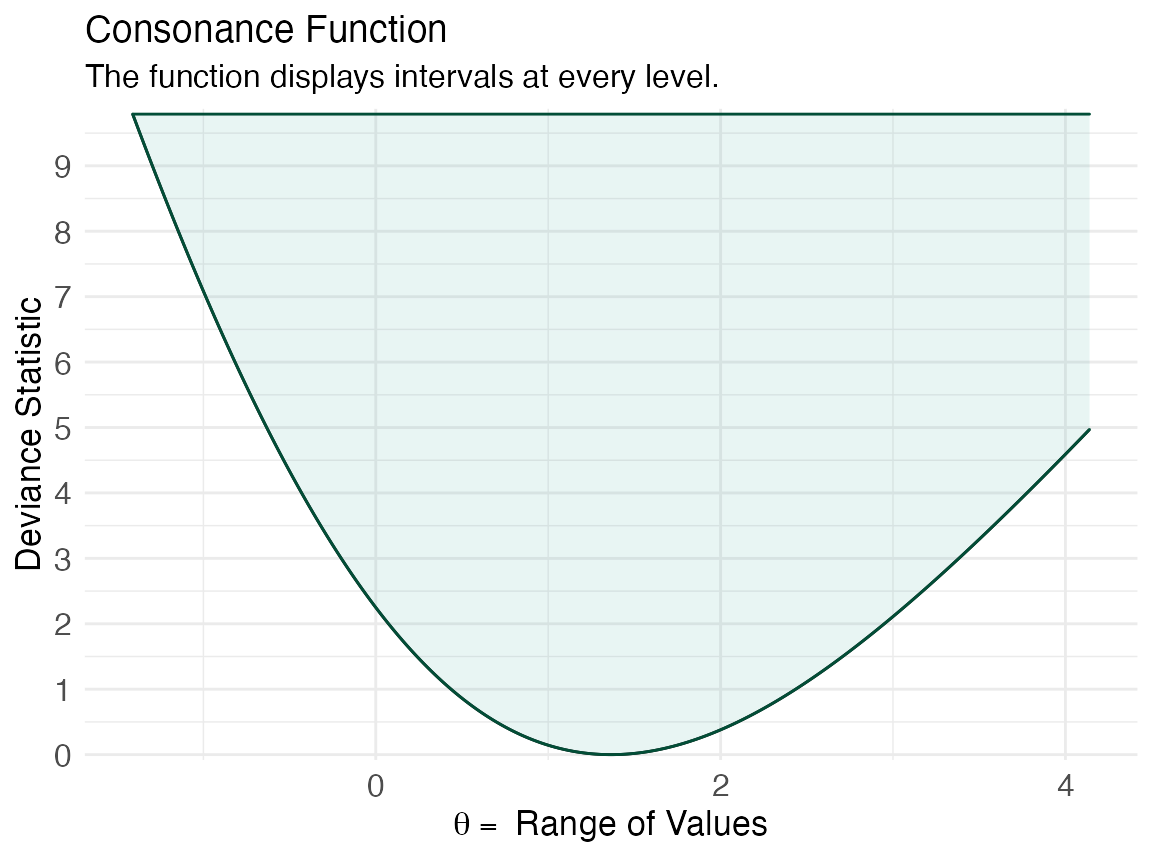
The obvious advantage of using reduced likelihoods is that they are free of nuisance parameters
\[L_{t_{n}}(\theta)=f_{n}\left(F_{n}^{-1}\left(H_{p i v}(\theta)\right)\right)\left|\frac{\partial}{\partial t} \psi\left(t_{n}, \theta\right)\right|=h_{p i v}(\theta)\left|\frac{\partial}{\partial t} \psi(t, \theta)\right| /\left.\left|\frac{\partial}{\partial \theta} \psi(t, \theta)\right|\right|_{t=t_{n}}\] thus, giving summaries of the data that can be incorporated into combined analyses.
Cite R Packages
Please remember to cite the packages that you use.
citation("concurve") #> #> Rafi Z, Vigotsky A (2020). _concurve: Computes and Plots Compatibility #> (Confidence) Intervals, P-Values, S-Values, & Likelihood Intervals to #> Form Consonance, Surprisal, & Likelihood Functions_. R package version #> 2.7.7, <URL: https://CRAN.R-project.org/package=concurve>. #> #> Rafi Z, Greenland S (2020). "Semantic and Cognitive Tools to Aid #> Statistical Science: Replace Confidence and Significance by #> Compatibility and Surprise." _BMC Medical Research Methodology_, *20*, #> 244. ISSN 1471-2288, doi: 10.1186/s12874-020-01105-9 (URL: #> https://doi.org/10.1186/s12874-020-01105-9), <URL: #> https://doi.org/10.1186/s12874-020-01105-9>. #> #> To see these entries in BibTeX format, use 'print(<citation>, #> bibtex=TRUE)', 'toBibtex(.)', or set #> 'options(citation.bibtex.max=999)'. citation("ProfileLikelihood") #> #> To cite package 'ProfileLikelihood' in publications use: #> #> Leena Choi (2011). ProfileLikelihood: Profile Likelihood for a #> Parameter in Commonly Used Statistical Models. R package version 1.1. #> https://CRAN.R-project.org/package=ProfileLikelihood #> #> A BibTeX entry for LaTeX users is #> #> @Manual{, #> title = {ProfileLikelihood: Profile Likelihood for a Parameter in Commonly Used Statistical #> Models}, #> author = {Leena Choi}, #> year = {2011}, #> note = {R package version 1.1}, #> url = {https://CRAN.R-project.org/package=ProfileLikelihood}, #> } #> #> ATTENTION: This citation information has been auto-generated from the #> package DESCRIPTION file and may need manual editing, see #> 'help("citation")'.
References
1. Choi L. ProfileLikelihood: Profile likelihood for a parameter in commonly used statistical models. 2011. https://CRAN.R-project.org/package=ProfileLikelihood.
2. Cole SR, Chu H, Greenland S. Maximum Likelihood, Profile Likelihood, and Penalized Likelihood: A Primer. American Journal of Epidemiology. 2013;179(2):252-260. doi:10/f5mx4q